Act I: Initiation
mRNA, a special type of RNA that is made in the process of protein-making is transcribed from a template of DNA in the nucleolus. First, to start the process, a group of proteins called the transcription factors attach themselves to the promoter region. The promoter region is a part of the gene sequence that marks where the gene begins, it is also known as upstream. In the promoter region is the TATA box, a genetic sequence that TFs bind to specifically. After the TFs have bonded to the promoter region, this signals another protein, RNA Polymerase II, to bind to the transcription factors. This entire group of protein creates what is called the transcription initiation complex.
Act II: Elongation
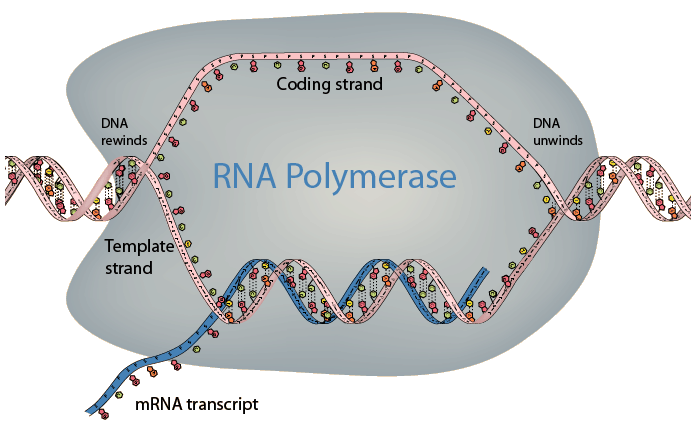
Polymerase II unwinds the double helix of the DNA, about 10 bases at a time and begins to synthesize pre-mRNA (AKA RNA transcript or primary RNA) Like always, new nucleotides are synthesized from 5' to 3'. During this synthesis, Polymerase II will begin to move downstream. Genetic information is only carried on one strand of the DNA, this strand is called the coding strand, also known as the sense strand. The coding strand is the strand that has the same genetic sequence as the newly synthesized pre-mRNA, with one exception: RNA does not have thymine, instead, it has a base called uracil. The template strand I (or antisense strand) is the complementary strand to the coding strand. Polymerase II uses the template strand to make complementary bases. The RNA tail becomes detached from the template strand as Poly II moves along.
Act III: Termination
As Polymerase II reaches the end of a gene, it will approach the terminator sequence which is a signal for Poly II to stop transcribing. In the terminator sequence, a sequence of "AAUAAA" will be found on the coding strand. Note that more than one Polymerase II can be working on a single gene at the same time.
As Polymerase II reaches the end of a gene, it will approach the terminator sequence which is a signal for Poly II to stop transcribing. In the terminator sequence, a sequence of "AAUAAA" will be found on the coding strand. Note that more than one Polymerase II can be working on a single gene at the same time.
Before pre-mRNA is sent out into the cytoplasm of the cell, several modifications need to happen. Because RNA is single stranded, it is unstable. The oxygen in RNA will react if it comes in contact with water. To combat this, a G-cap is added to the 5' end of RNA and a poly-A-tail (many adenines) is added onto the 3' end of RNA. As well as protection, the G-cap also acts as an "attach here" signal for ribosomes. The poly-A-tail helps in the export of the RNA into the cytoplasm.
Since there is no proof-reading, as there is in DNA synthesis, nature has implemented a few methods of ensuring there are a few mistakes as possible. The first method is called RNA splicing. In this process, "junk sequences," called introns are removed from the RNA. What is left after are only the exon sequences. The splicing is done by a spliceosome, a variety of proteins, including a few snRNPs (small nuclear ribonuclear proteins). Inside the snRNPs are snRNAs (small nuclear RNA molecules). These snRNAs recognizes intron RNA and bonds with it and then cuts the transcript RNA to release the intron and exons are connected together.
If any mutations occurred in the introns, they will have no effect on the final protein, since it is spliced away.


No comments:
Post a Comment